I am a Ph.D Candidate in Biology (Computional Biology) at the School of Life Sciences, Westlake University, advised by Prof Weike Pei & Prof Yanxiao Zhang. My research focuses on decoding cell fate decision processes by integrating single-cell and spatial genomics with machine learning. Before joining Westlake, I developed algorithms for single-cell multi-omics data analysis and network inference at Jie Wang’s lab in GIBH-CAS.
I am also an amateur bodybuilder with 135KG maximal weight for bench press, 190KG maximal weight for Deadlift, and 170KG maximal weight for Squat.
🔈 Seeking Postdoc Opportunities
- I am actively seeking a postdoctoral position in computational single-cell and spatial genomics outside mainland China. I would greatly appreciate it if you could share any relevant opportunities (Please email me at jiangjunyao@westlake.edu.cn).
🔥 Summary of Main Research Topic
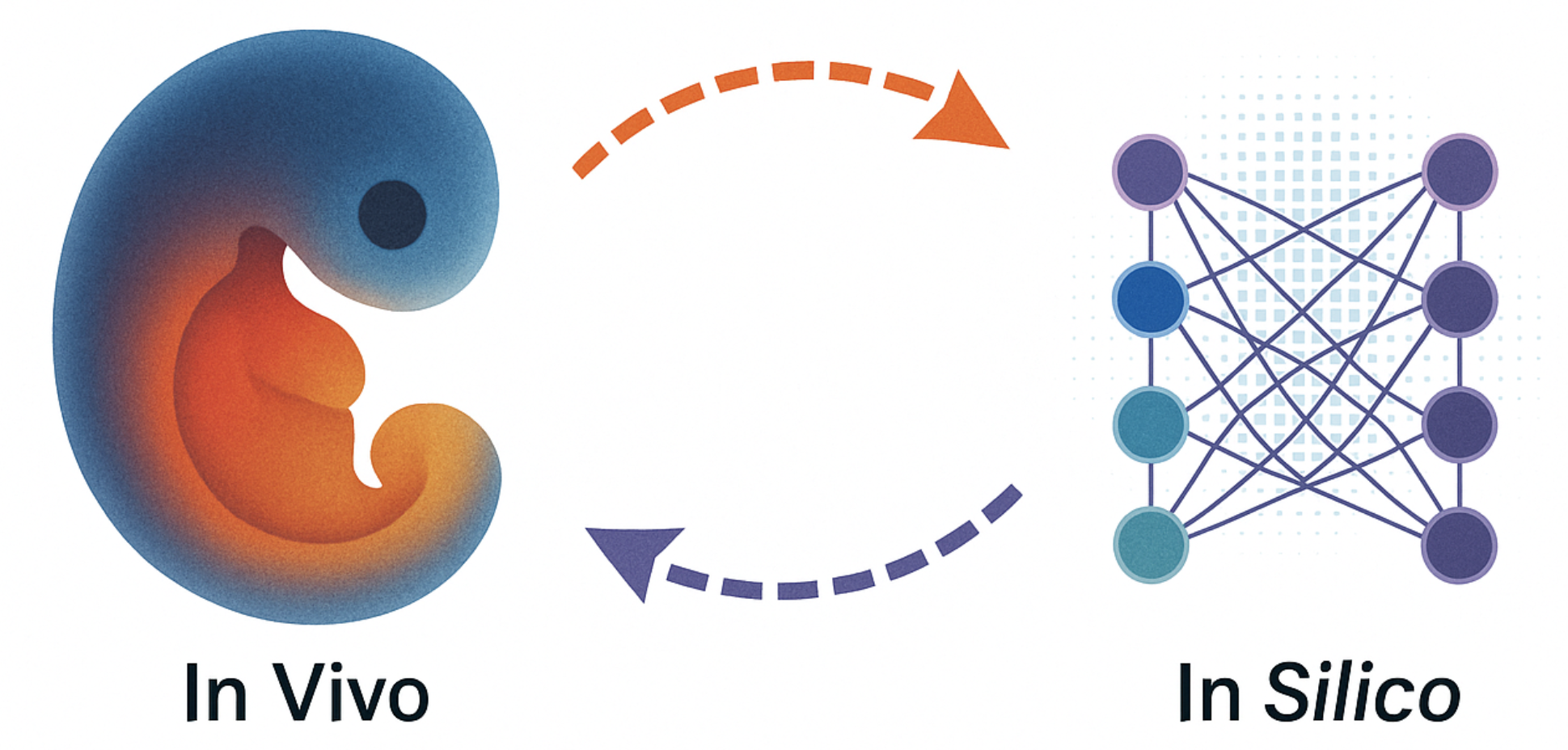
- Cell fate decisions are crucial for understanding how pluripotent cells develop into diverse tissues, with disruptions leading to congenital abnormalities.
- Through in vivo single-cell multiomics lineage tracing, such as DeepTrack barcoding, we have uncovered early lineage priming in the mouse embryo and reconstructed fate-resolved GRNs, identifying key regulator of mesoderm development.
- However, direct genetic barcoding is ethically and technically unfeasible in human embryos, necessitating the development of non-invasive approaches. To address this challenge, we have developed transfer learning-based methods for in silico fate mapping, enabling the discovery of spatiotemporal fate biases through data derived from 3D human embryo.
📝 Publications
†: co-first, #: correspondence
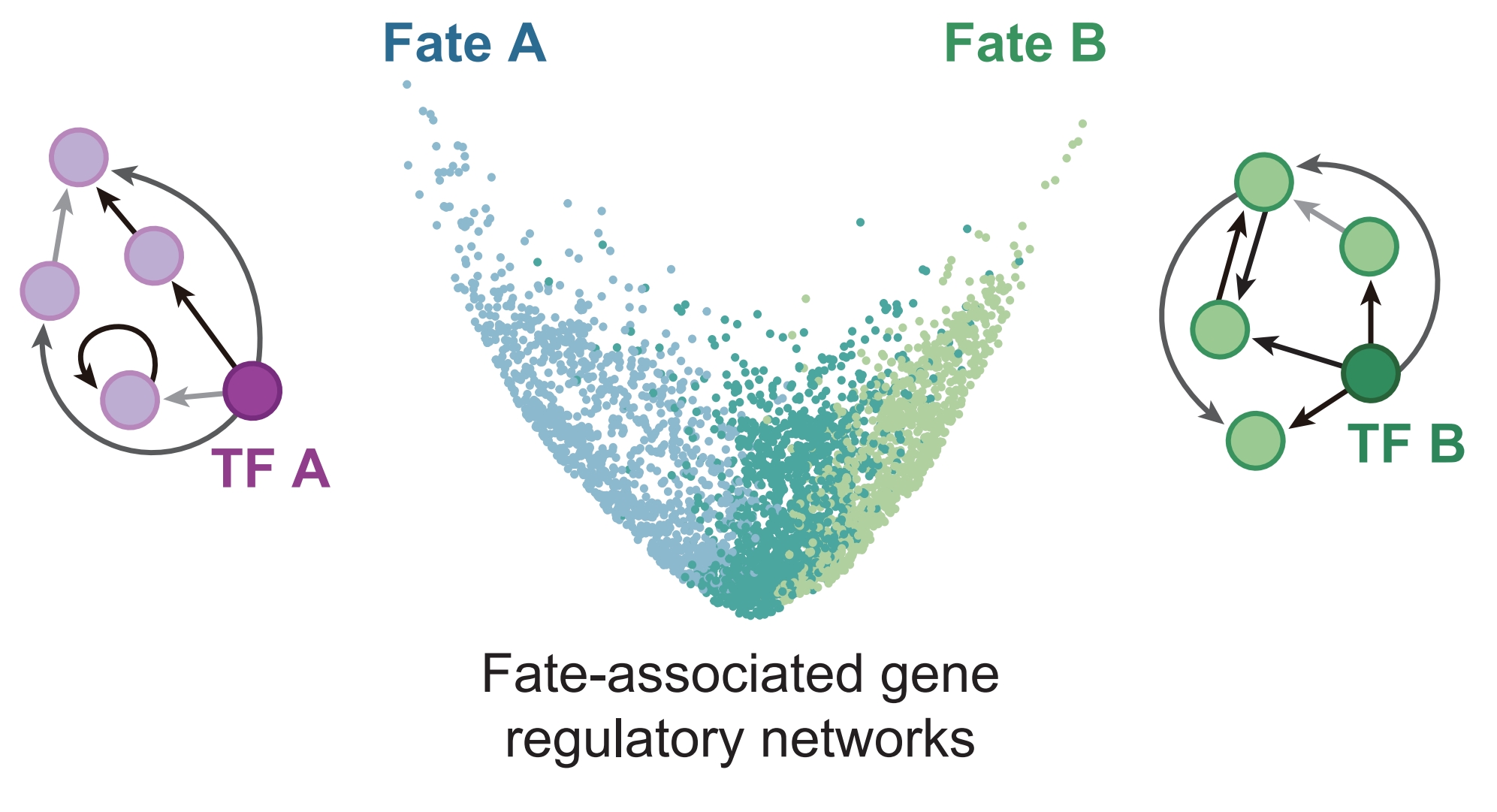
In vivo multimodal lineage tracing of mammalian development by DeepTrack barcoding
Chenyu Guo†, Junyao Jiang†, Xiaomin Wang†, Xinghuai Huang, Shenghu Zhang, Chenyang Shao, Mingyuan Zhang, Xiwen Hu, Wenqian Yang, Fuwei Shang, Xi Wang, Hongbo Zhai, Quan Du, Fang Liu, Danyang He, Xiaodong Liu, Guangdun Peng, Saifeng Cheng, Yanxiao Zhang#, Duanqing Pei#, Weike Pei#
Keywords: Single-cell multi-omics Lineage tracing, Mouse embryogenesis, Fate resolved GRN, Cell atlas
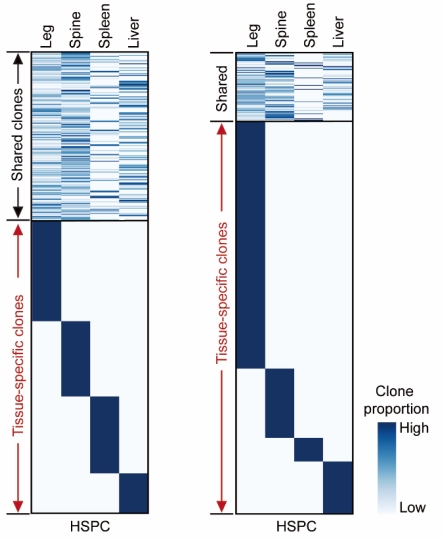
Uncovering the origins and functional features of hematopoiesis in extramedullary organs
Wenqian Yang†, Zhexuan Hu† , *Junyao Jiang†*, Chenyu Guo†, Zongcheng Li†, Weipeng Ge, Yizhen Chu, Qi Shi, Xiaotong Wang, Xinghuai Huang, Xiwen Hu, Hui Li, Huayu Pan, Fuwei Shang, Zheng Wang, Xi Wang, Fang Liu, Quan Du, Hongbo Zhai, Heping Xu, Danyang He, Yu Lan#, Bing Liu#, Weike Pei#
Keywords: Lineage tracing, Extramedullary hematopoiesis, EPO
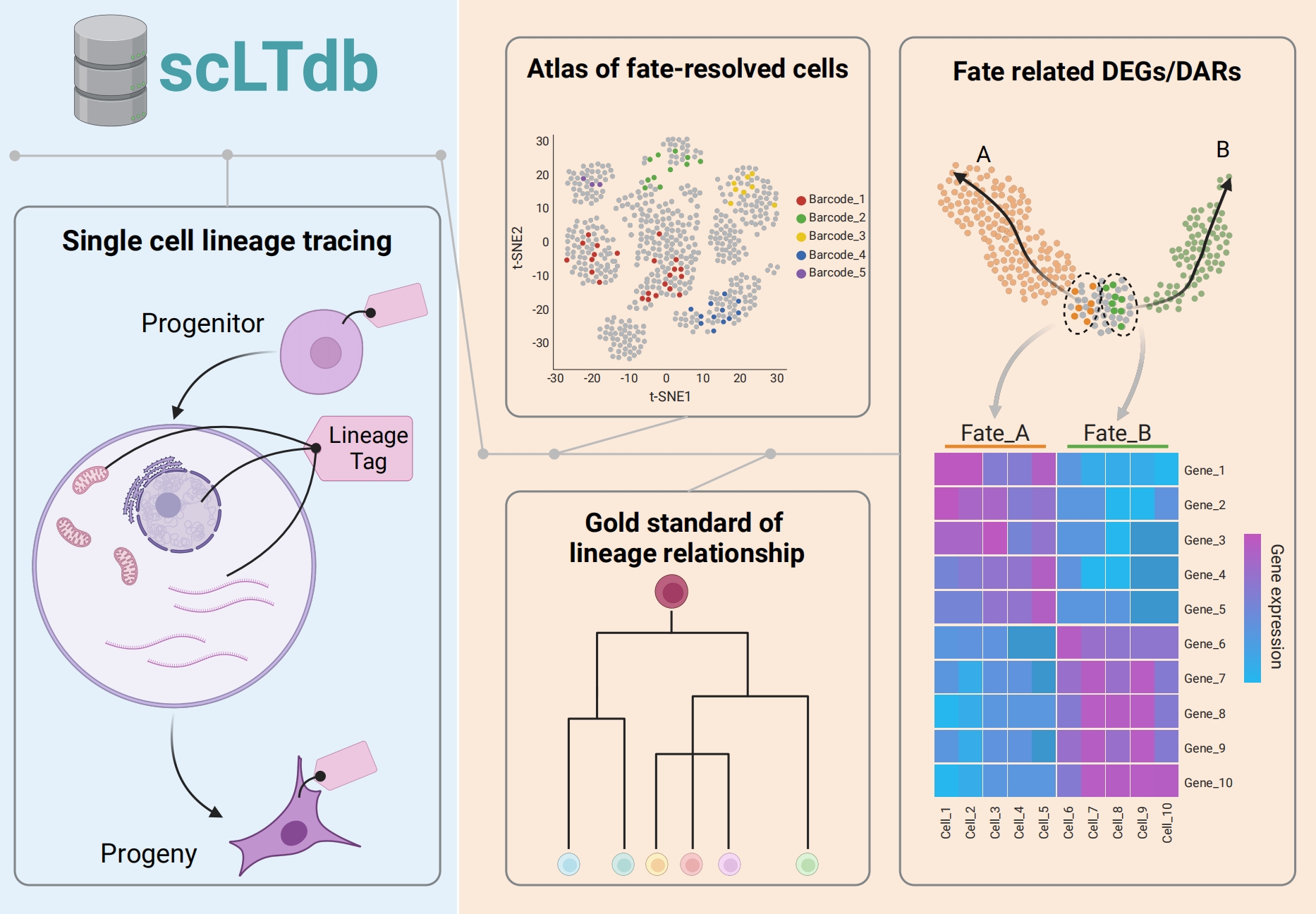
scLTdb: a comprehensive single cell lineage tracing database
Junyao Jiang†, Xing Ye†, Yunhui Kong†, Chenyu Guo, Mingyuan Zhang, Fang Cao, Yanxiao Zhang#, Weike Pei#
Keywords: Single-cell and spatial genomics, Lineage tracing, Online resource
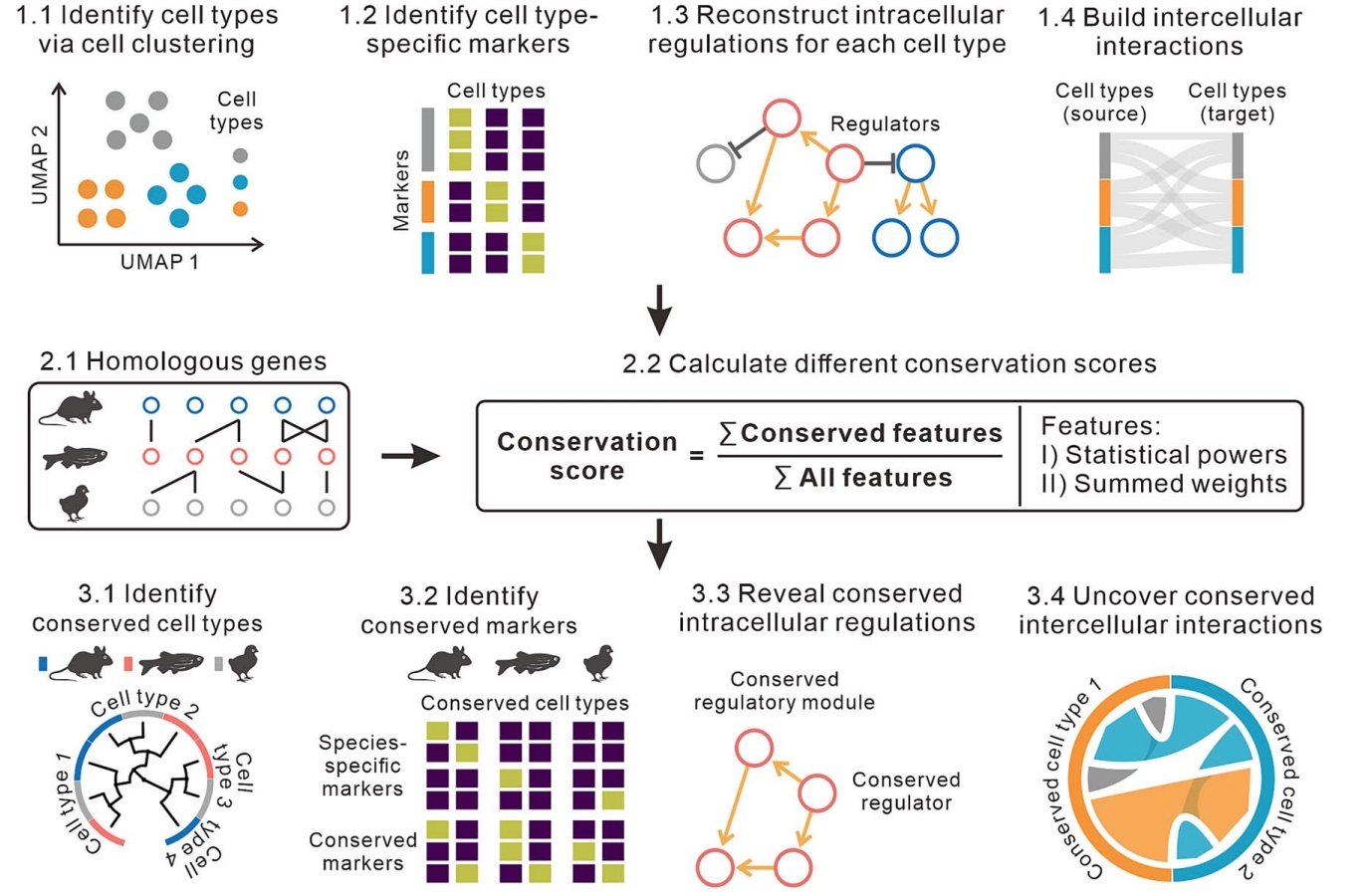
Junyao Jiang†, Jinlian Li†, Sunan Huang, Fan Jiang, Yanran Liang, Xueli Xu#, Jie Wang#
Keywords: scRNA, Cross-species comparison, Conserved regulatory programs
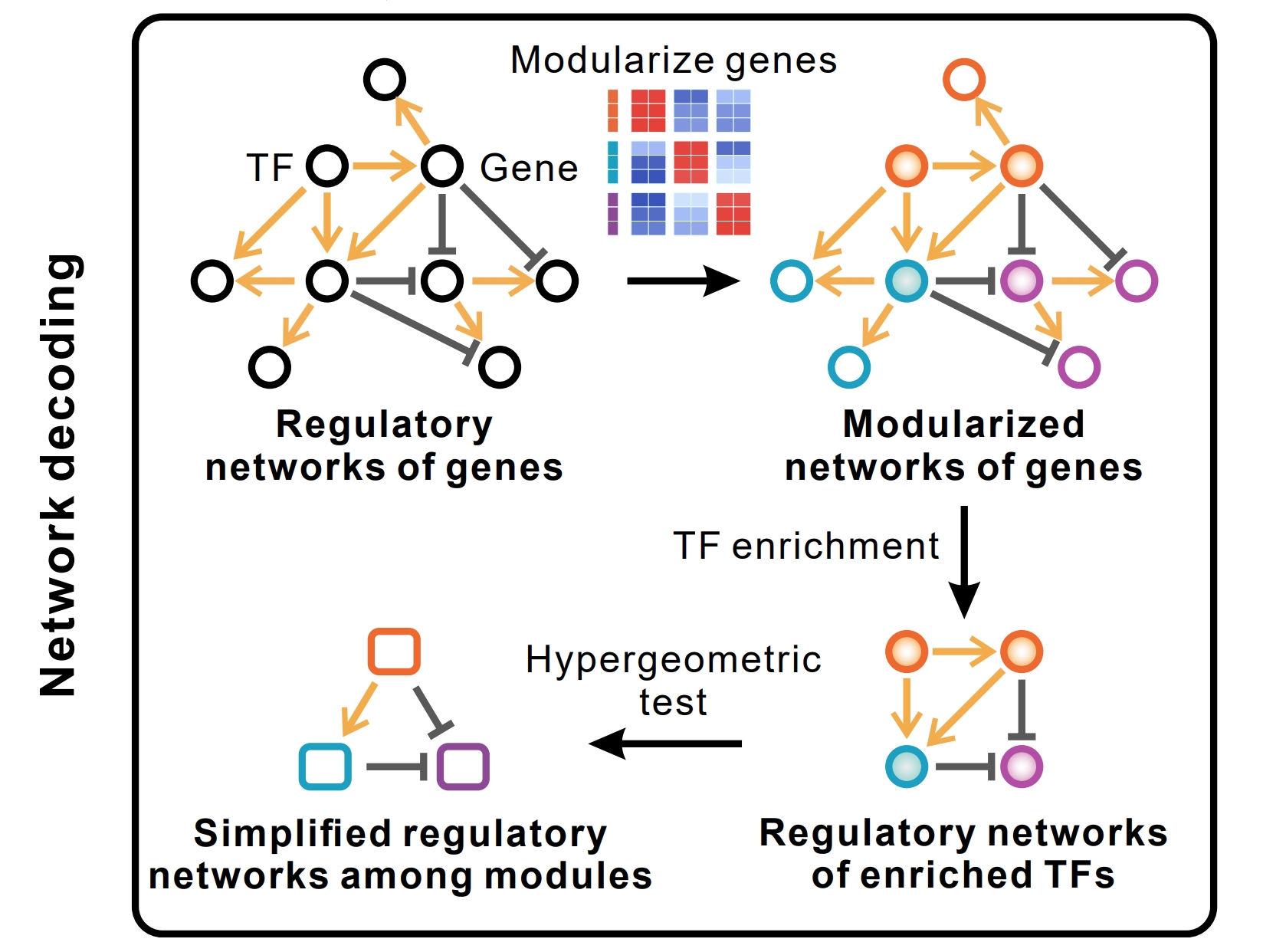
Junyao Jiang†, Pin Lyu†, Sunan Huang, Jiawang Tao, Seth Blackshaw, Qian Jiang, Jie Wang
Keywords: Single-cell multiomics, Gene regulatory netowrk, Inter celltype regulations
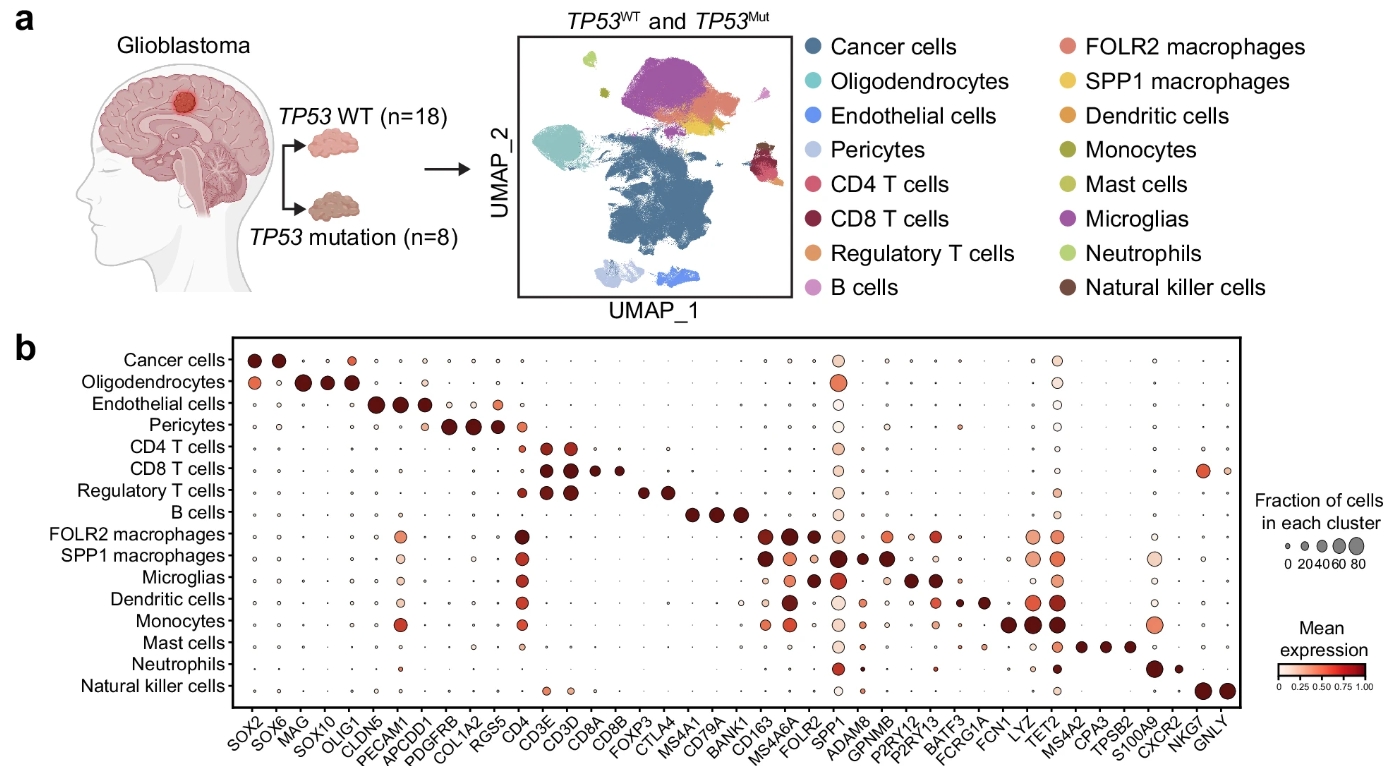
Yapeng Ji, Junyao Jiang, Lei Hu, Peng Lin, Mingshan Zhou, Song Hu, Minkai Wang, Yuchen Ji, Xianzhi Liu, Dongming Yan, Yang Guo, Adwait Amod Sathe, Bret M. Evers, Chao Xing, Xuelian Luo, Qi Xie, Weike Pei, Zhenyu Zhang#, Hongtao Yu#
Keywords: scRNA, Cell atlas, Glioblastoma, Cancer immunotherapy
- Yunhui Kong†, Junyao Jiang†,#, Weikang Kong, Sheng Qin#. DRCTdb: disease-related cell type analysis to decode cell type effect and underlying regulatory mechanisms. Communications Biology, Sep 2024 (Co-first & Co-correspondence)
- Ying Xin, Pin Lyu, Junyao Jiang, Fengquan Zhou, Jie Wang, Seth Blackshaw, Jiang Qian. LRLoop: Feedback loops as a design principle of cell-cell communication. Bioinformatics, July 2022
- Dapeng Sun†, Xiaojie Gan†, Lei Liu†, Yuan Yang†, Dongyang Ding, Wen Li, Junyao Jiang, et al. DNA hypermethylation modification promotes the development of hepatocellular carcinoma by depressing the tumor suppressor gene ZNF334. Cell Death & Disease, May 2022
🎖 Awards
- 2024.10 Doctoral National Scholarship (Top 1% PhD student in China, ~4500 USD)
🚩 Professional Experience in Computional Biology
- 2022.08 - 2026.06, Ph.D Candidate in Biology (Computional Biology) at Westlake University
- 2021.03 - 2022.06, Research Assistant at Guangzhou Institutes of Biomedicine and Health, Chinese Academy of Sciences
- 2020.12 - 2021.02, Bioinformatician Intern at Singleron Biotech
- 2020.12 - 2021.02, Master of Science in Genomics & Bioinformatics at Chinese University of Hong Kong
📛 Mentorship
- Xing Ye: Undergraduate from University of Science and Technology of China, 2023.11-2024.06. (Co-first author at scLTdb project)
- Zhi Chen: Undergraduate from Tsinghua University, 2025.07-2025.08. (Summer research programe)
- Jiayi Liu: Research Assistant from Imperial College London, 2025.10-Present.
📞 Peer Review Service
- Nature (Co-Review)
- Communications Biology
- Archives of Computational Methods in Engineering
- BMC Bioinformatics
- BMC Genomics
- Scientific Reports
💻 Maintained Software
- IReNA Consturct gene regualtory networks based on single cell multiomics data
- CACIMAR scRNA-seq based cross-species data analysis
- FateMapper single cell lineage tracing data analysis and visualization
- scLTdb Database for single cell lineage tracing
- FateExplorer Machine learning method to generate clone embedding and perform fate analysis
